
Pharmacogenomics: Increasing the safety and effectiveness of drug therapy
Pharmacogenomics
One of the most important components of personalized medicine is pharmacogenomics, the study of genetic variations that influence individual response to drugs. Enzymes responsible for drug metabolism and proteins that determine the cellular response to drugs (receptors) are encoded by genes, and can therefore be variable in expression, activity level and function when genetic variations are present. Knowing whether a patient carries any of these variations may help health care professionals individualize drug therapy, decrease the number of adverse drug reactions and increase the effectiveness of drugs. Pharmacogenomics has been characterized as “getting the right dose of the right drug to the right patient at the right time.”2 This brochure is intended to introduce the concept of pharmacogenomics to physicians and other health care providers using a case-based approach. Note that the terms “pharmacogenomics” and “pharmacogenetics” are often used interchangeably; for this brochure, “pharmacogenomics” will be used.
Genes commonly involved in pharmacogenomic drug metabolism and response
There are several genes responsible for differences in drug metabolism and response. Among the most common are the Cytochrome P450 (CYP) genes, encoding enzymes that control the metabolism of more than 70 percent of prescription drugs. People who carry variations in certain CYP genes often do not metabolize drugs at the same rate or extent as in most people, and this can influence response in many ways. Other genes known to affect drug response encode the receptors for regulatory molecules such as neurotransmitters, hormones, cytokines and growth factors, and cellular proteins such as enzymes, transporters, carriers, ion channels, structural proteins and transcription factors. Variations in these genes can lead to poor response and adverse drug reactions by disabling, inactivating, interfering with, or inaccurately inducing the signaling mechanisms or cellular machinery that must function for the body to respond properly to the drug; or by causing side effects that prevent continued use of the drug.
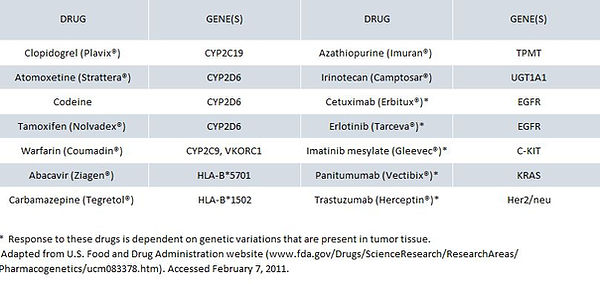
Selected drugs whose safety and efficacy are affected by gene variations
The table below is a partial list of drugs that exhibit reduced therapeutic effectiveness and/or safety concerns in patients carrying certain genetic variations. These variations often make the drug unsafe or unsuitable for patients who carry the variations. This list contains examples of drugs that are affected by inherited genetic variations and by variations that are acquired and present in tumor tissue.
Metabolizer phenotype
The metabolizer phenotype describes the patient’s ability to metabolize certain drugs and is based on the number and type of functional alleles of certain genes that a patient carries. These genes most commonly encode the CYP enzymes, which are the focus of much of this brochure. The metabolizer phenotype can range from “poor,” used to describe patients with little or no functional activity of a selected CYP enzyme, to “ultra-rapid,” used to describe patients with substantially increased activity of a selected CYP enzyme. Depending on the type of CYP variation present, the patient’s metabolizer phenotype and the type of drug (active pharmacologic agent or inactive prodrug precursor), therapeutic drug response is often suboptimal. The table on the following page summarizes the effects of CYP variation on therapeutic efficacy. For example, poor metabolizers are unable to metabolize certain drugs efficiently, resulting in a potentially toxic build-up of an active drug or the lack of conversion of a prodrug into an active metabolite. In contrast, in ultra-rapid metabolizers, an active drug is inactivated quickly, leading to a subtherapeutic response, while a prodrug is quickly metabolized, leading to rapid onset of therapeutic effect.

Some drugs and foods cause altered metabolizer phenotype
Certain drugs mimic the effect of genetic variations, effectively causing changes in metabolizer phenotype. For example, quinidine is an inhibitor of CYP2D6 activity. A patient taking quinidine is therefore a CYP2D6 poor metabolizer, similar to someone who carries a loss-of function variation in CYP2D6. In those patients, drugs that require the activity of CYP2D6, such as atomoxetine, will not be metabolized at the same rate as in most people.4 Certain foods can also mimic the effects of genetic variations. One of the most common examples is grapefruit juice, which is an inhibitor of CYP3A4. In people regularly drinking grapefruit juice, drugs that require the activity of CYP3A4, such as diazepam, will not be metabolized at the same rate as in most people.
Pharmacogenomics in the clinical setting
Awareness of the influence of gene variations on patient response to certain drugs can help physicians decide which type of drug therapy may be appropriate, and identify cases in which a patient isn’t responding as anticipated to a drug. The examples that follow illustrate three categories for which pharmacogenomic knowledge can help inform therapeutic decisions: predicting and preventing adverse reactions, determining the efficacy of a drug for a particular patient, and predicting the optimal drug dose.
Using pharmacogenomics to predict and prevent adverse drug reactions
Several drugs can cause severe or life-threatening reactions in patients with variations in genes that encode proteins that metabolize or are targets of the drugs. Knowing about patients’ genetic variations can help physicians avoid drugs that may cause adverse reactions. On the following two pages are examples of drugs in this category.
Abacavir
Abacavir (Ziagen®) is a nucleoside reverse transcriptase inhibitor used in combination with other antiretrovirals to treat HIV infection.5 An immunologically-mediated hypersensitivity reaction occurs in 5–8 percent of patients taking abacavir, usually during the first six weeks after initiation of therapy. The hypersensitivity symptoms include a combination of fever, rash, gastrointestinal tract symptoms and respiratory symptoms that become more severe with continued dosing.6 Discontinuation of abacavir therapy results in reversal of symptoms. The abacavir hypersensitivity described above is associated with a variant allele of the major histocompatibility complex, HLA-B*5701. Patients who carry the HLA-B*5701 allele have an increased risk for developing a hypersensitivity reaction.6 HLA-B*5701 screening before abacavir treatment results in a significantly reduced number of hypersensitivity cases.6
The abacavir product labeling recommends genetic testing to detect the presence of HLA-B*5701.5 In the “Warnings and Precautions” section, the labeling states:

Codeine
Codeine is a prodrug with analgesic properties due primarily to its conversion into morphine.7,8 Conversion to morphine is mediated by the cytochrome P450 enzyme CYP2D6. Variations that decrease the metabolic activity of CYP2D6 result in a poor analgesic response due to the reduced conversion of codeine into morphine,3 and patients carrying such a variation are considered poor metabolizers and receive little therapeutic benefit from codeine. It is estimated that 5–10 percent of Caucasians are CYP2D6 poor metabolizers; the percentage is approximately 2–3 percent in other racial and ethnic groups.7,9,10
Variations (such as gene duplications) can also result in increased metabolic activity of CYP2D6; these result in an enhanced analgesic response due to the rapid conversion of codeine into morphine. Patients who carry such variations are at risk for opioid toxicity, which includes moderate to severe central nervous system depression. The prevalence of the CYP2D6 ultra-rapid metabolizer phenotype has been estimated at 1–10 percent in Caucasians, 3–5 percent in African Americans, 16–28 percent in North Africans, Ethiopians and Arabs, and up to 21 percent in Asians.3,11
The product labeling (updated in July 2009) of drugs containing codeine include warnings that CYP2D6 ultra-rapid metabolizers “may experience overdose symptoms such as extreme sleepiness, confusion or shallow breathing, even at labeled dosage regimens,” and encourages physicians to “choose the lowest effective dose for the shortest period of time and inform their patients about the risks and the signs of morphine overdose.”11 Of additional concern is the use of codeine in nursing mothers. Product labeling includes the statement that women who are CYP2D6 ultra-rapid metabolizers achieve “higher-than-expected levels of morphine in breast milk and potentially dangerously high serum morphine levels in their breastfed infants.” A 2007 U.S. Food and Drug Administration (FDA) Public Health Advisory warns about the dangers to neonates of codeine use in breast-feeding mothers and states that the only way to determine prior to drug administration whether a patient is an ultra-rapid metabolizer is by the use of a genetic test.12 Canadian guidelines also state that genetic testing is available.
Case study
DS is a 30 year-old woman who gave birth by caesarian section 10 days ago. Her physician prescribed codeine for post-caesarian pain. Despite taking no more than the prescribed dose, DS experienced nausea and dizziness while she was taking codeine. She also noticed that her breastfed infant was lethargic and feeding poorly. When DS mentioned these symptoms to her physician, he recommended that she discontinue codeine use. Within a few days, both DS’s and her infant’s symptoms were no longer present.
How would genetic testing help DS and her physician? Genotyping of DS’s CYP2D6 gene may have revealed a duplication of CYP2D6 genes, placing her in the ultra-rapid metabolizer category. Armed with this knowledge, DS’s physician would likely have prescribed a different analgesic that would have spared DS and her infant the symptoms of opioid toxicity.
Using pharmacogenomics to predict effectiveness
Several drugs are subtherapeutic or ineffective in patients with variations in genes that encode drug-metabolizing proteins or targets of the drugs. Knowing about patients’ genetic variations can help physicians select drug therapies that will be most effective for individual patients. Below is an example of a drug in this category.
Clopidogrel
Clopidogrel (Plavix®) is a platelet inhibitor used in the treatment of a number of cardiovascular diseases. It is often prescribed for secondary prevention following acute coronary syndromes and for those undergoing percutaneous coronary intervention. However, despite Clopidogrel treatment, up to one-quarter of patients experience a subtherapeutic antiplatelet response, resulting in a higher risk for ischemic events.14,15
Clopidogrel is a prodrug; its antiplatelet properties are exerted once it is converted to an active metabolite.16 A cytochrome P450 enzyme, CYP2C19, mediates the conversion of cCopidogrel into the active metabolite. Patients who carry certain variations in CYP2C19 are considered poor metabolizers and show reduced ability to convert Clopidogrel into its active metabolite, resulting in a diminished antiplatelet effect.16,17 Further, these patients are more likely to have an ischemic event following Copidogrel therapy.17 Approximately 2–20 percent of patients (depending on ethnicity) are likely to carry CYP2C19 variations.3,18
A Boxed Warning is included in the product labeling to alert health care providers about its reduced effectiveness in patients who are CYP2C19 poor metabolizers, and to inform health care providers that genetic tests are available to detect genetic variations in CYP2C1918:
Case study
JM is a 58 year-old man who recently had an acute myocardial infarction. To prevent subsequent ischemic events, JM’s physician recommends antiplatelet therapy and prescribes Clopidogrel. Six months later, JM suffered another acute MI, and his physician suspects that the patient has been non-adherent, or alternatively, that Clopidogrel therapy may have been ineffective.
How would genetic testing help JM and his physician? Determining JM’s CYP2C19 genotype may reveal that he carries a variant that diminishes the antiplatelet effect of Clopidogrel. If this were the case, alternative anti-platelet therapies may have been considered, reducing the chance that JM would suffer a second cardiac event.
In patients taking both Clopidogrel and the proton pump inhibitors omeprazole or esomeprazole, diminished antiplatelet activity and adverse cardiac outcomes have been observed.19 It is thought that omeprazole and esomeprazole inhibit the activity of CYP2C19, making patients who take these drugs de facto CYP2C19 poor metabolizers, similar to someone who carries a loss-of function variation in CYP2C19. In these patients, drugs that require the activity of CYPC19, such as Clopidogrel, will not be metabolized at the same rate as in most people.
Using pharmacogenomics to predict optimal dose
Genetic variations can lead to an altered dosage regimen. Knowing whether a patient carries these genetic variations can assist physicians in determining optimal therapeutic dose. An example of a drug with an altered dosage regimen in patients with certain genetic variations is warfarin.
Warfarin
Warfarin (Coumadin®) is the most widely-prescribed anticoagulant used to treat and prevent thromboembolic diseases. It is metabolized by the CYP2C9 enzyme, and its anticoagulant effect is mediated by the enzyme VKORC1. Variation in the CYP2C9 gene causes some patients to have slow metabolism of warfarin and a longer half-life of the drug, resulting in higher than usual blood concentrations of warfarin and greater anticoagulant effect. Certain variations in the VKORC1 gene result in reduced activity of the enzyme and subsequently reduced synthesis of coagulation factors. The combination of slow warfarin metabolism caused by CYP2C9 gene variations and reduced coagulation caused by VKORC1 gene variations increases the risk of bleeding during warfarin therapy.
Warfarin has a narrow therapeutic index; variations in CYP2C9 and VKORC1, in addition to several other patient characteristics, make it difficult to predict the effective dose. Those carrying certain CYP2C9 and VKORC1 variations are likely to require altered doses and may require prolonged time to reach a stable maintenance dose. The prevalence of these CYP2C9 and VKORC1 variations is variable among racial and ethnic groups: up to 17 percent of Caucasians, 4 percent of African-Americans and less than 2 percent of Asians carry at least one variant of CYP2C9;20 while 37 percent of Caucasians, 14 percent of African-Americans, and 89 percent of Asians carry at least one variant in VKORC1.21
9
The warfarin product labeling (updated January 2010) states “The patient’s CYP2C9 and VKORC1 genotype information, when available, can assist in selection of the starting dose,” and includes the following table of expected therapeutic doses for patients with different combinations of CYP2C9 and VKORC1 variations:22
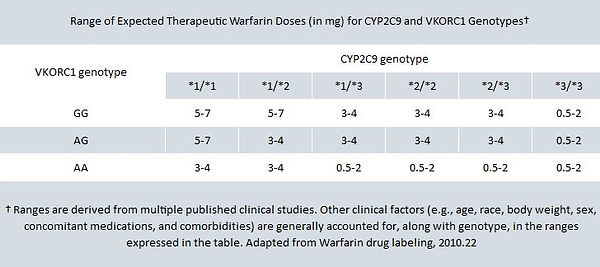
The product labeling suggests that this table of expected therapeutic doses can be used to assist prescribers in choosing initial warfarin dose for patients whose CYP2C9 and VKORC1 genotypes are known. For example, if genetic testing reveals that a patient carries the *1/*3 variation of CYP2C9 and the AG variation of VKORC1, his or her expected therapeutic warfarin dose is likely to be in the 3–4 mg range.
Dosing algorithms that rely on clinical features such as age, sex and weight, along with genotype, can also assist in the determination of optimal dose (visit www.warfarindosing.org, where one such algorithm can be found). Genotype is one factor of many that contribute to variation in a patient’s response to warfarin. Careful monitoring of INR is still required during titration to steady state and monitoring of long-term therapy.
Case study
ML is a 65 year-old woman who has recently been diagnosed with atrial fibrillation. To reduce the risk of stroke and other thrombotic events, ML’s physician recommends warfarin therapy. To estimate the initial dose, ML’s clinical characteristics (age, sex, weight, diet) were considered. However, ML will need to return to the clinic every day for INR monitoring until a stable dose is determined, and then every few weeks thereafter for maintenance monitoring.
How would genetic testing help ML and her physician? Determination of ML’s CYP2C9 and VKORC1 genotype would reveal whether she carries any variations that alter her ability to metabolize and respond to warfarin. Knowing about any gene variations before initiating therapy allows for more accurate initial dosing and faster INR stabilization, and can reduce the risk for bleeding or clotting events.
Pharmacogenomics and genetic testing
Genetic tests that detect variations in the genes that control metabolism and response to certain drugs are commercially available. Costs and insurance coverage are variable, depending on the type of test ordered. Your hospital and reference laboratories as well as clinical geneticists are valuable sources of information on the best test to order and interpretation of results.
It is important to note that the field of pharmacogenomics is still developing, with new findings being rapidly reported. Clinical trials and other studies focusing on the benefits, risks and cost-effectiveness of using genetic information to inform drug therapy are underway. In the meantime, physicians and health care providers should be familiar with the concept that genetic variations can cause their patients to respond unexpectedly to drug therapy, and that in some cases, it may be appropriate to use genetic testing to guide therapeutic decisions. For more information on pharmacogenomics, visit the following websites:
• UC San Diego Skaggs School of Pharmacy and Pharmaceutical Sciences Pharmacogenomics Education Program (http://pharmacogenomics.ucsd.edu/home.aspx)
• American College of Clinical Pharmacology The Future of Medicine: Pharmacogenomics (http://user.accp1.org/Sample_Home.htm)
• Indiana University School of Medicine, Division of Clinical Pharmacology Defining Genetic Influences on Pharmacologic Responses (www.drug-interactions.com)
• U.S. Food and Drug Administration Genomics (www.fda.gov/Drugs/ScienceResearch/ Research Areas/Pharmacogenetics/default.htm)
• Pharmacogenomics Research Network (www.nigms.nih.gov/initatives/pgrn)
Acknowledgements The American Medical Association, the Arizona Center for Education and Research on Therapeutics (AzCERT), and the Critical Path Institute thank the American Academy of Family Physicians, the American College of Clinical Pharmacy, the American Society for Clinical Pharmacology and Therapeutics, Medco Research Institute, the Personalized Medicine Coalition, and several individuals with expertise in pharmacogenomics for their helpful review comments. This brochure was partially funded by a grant from the Agency for Healthcare Research and Quality to AzCERT. The authors of this document declare no conflicts of interest. The information contained in this document is not intended to be a treatment recommendation or an endorsement of any particular product..